TE-derived TSS Region: chr10:100381166..100381267
This page displays the conservation of transposon (TE) - derived transcription start site (TSS) region across mammals.
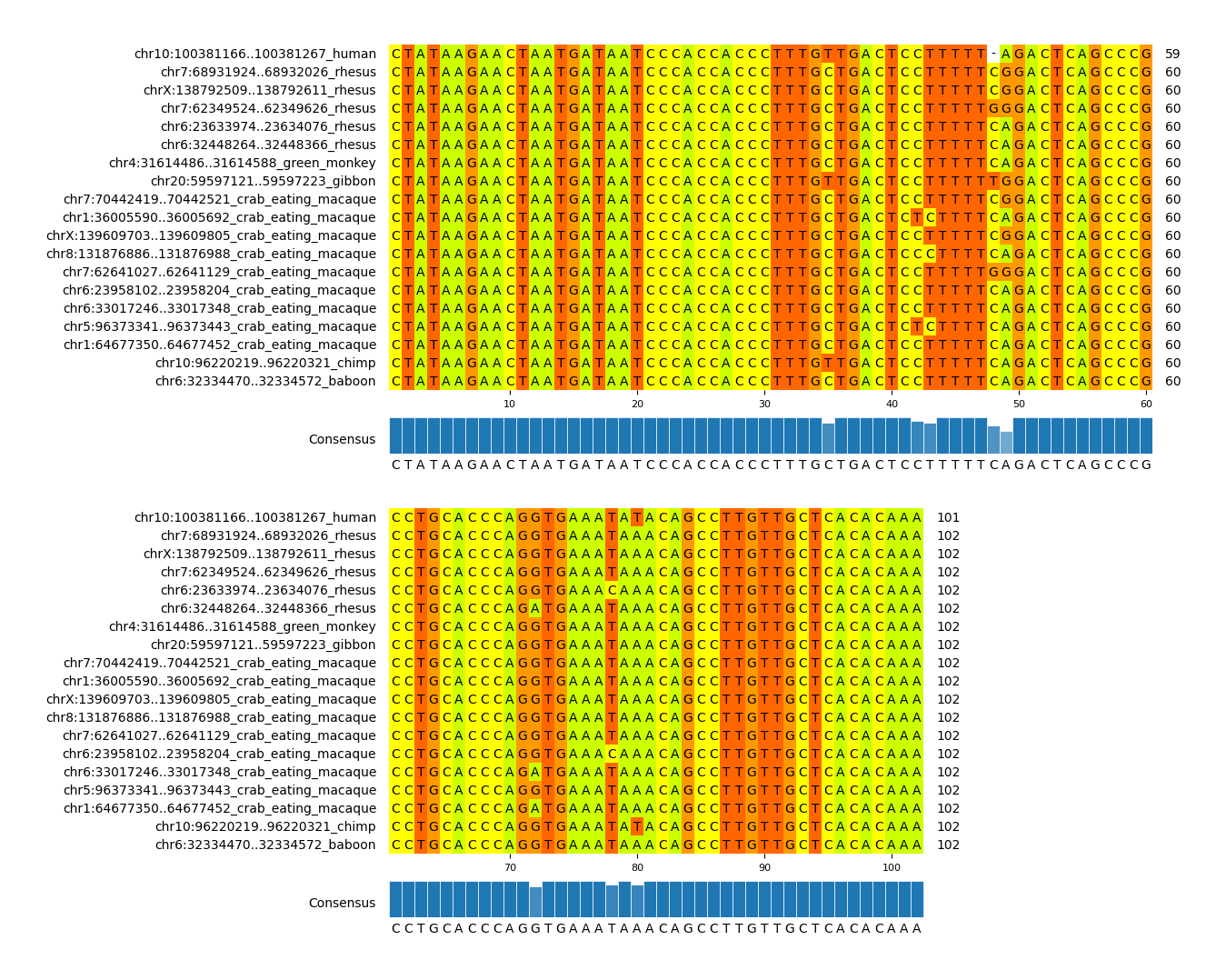
This figure displays the multiple sequence alignment of human TE-derived TSS region and homologous regions in mammals. You can discern the sequence similarity between them.
This figure illustrates the modified BLAT score of human TE-derived TSS region compared to other mammals based on BLAT analysis. To ensure alignment accuracy, we have selectively retained sequences with a modified BLAT score greater than 0.7.
Through the genome browser, you can explore the sequence features of homologous regions, including the presence of TE elements and their involvement as TSSs.
Copyright © 2023.Tongji University, Life Science and Technology Department, Xiao-Ou Zhang Lab
Contact us: zhangxiaoou@tongji.edu.cn
Related Resource: Zhang-Lab Website